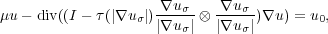
In biology, mutant strain analysis is an important tool for understanding the lipid metabolism of the yeast Saccharomyces Cerevisiae. For that purpose, 3D fluorescence laser microscopy and 2D transmission light microscopy images of yeast cell colonies are examined. The goal is to obtain cell-based quantitative information about the lipid droplets from the fluorescence images. However, the necessary information about the cell boundaries is only present in the 2D transmission images which exhibit a strongly heterogeneous structure. This causes standard thresholding-based methods to fail. The objective is to develop a cell segmentation strategy which is suitable for this class of images.
A data-processing pipeline performing the segmentation has been proposed and implemented. First, to provide robustness for the subsequent steps, the transmission image u0 is preprocessed with the non-linear degenerate elliptic smoothing
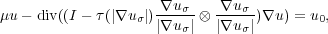
which suitable homogeneous Neumann boundary conditions, μ > 0, uσ = u * Kσ and a suitable τ : [0,∞[ → [0,1].
The heart of the algorithm is a modified active contour model which is used for the segmentation of a single cell. For the smoothed image u, it carries out numerical optimization for the problem
![∫ α ∫ ′ 2 β ∫ ′ ⊥ p
γm:Si1n→R2 S1 G(γ(t))dt+ 2- S1 |γ(t)| dt- (2 S1 γ(t)⋅γ (t) dt) ,
2
G (x) = (1+ |∇u(x2)|-)- 1, λ > 0, α,β > 0, p ∈ ]0,1[.
λ](snakes1x.png)
The GUI-based implementation was tested with 523 data sets taken from various yeast mutant colonies. Visual inspection shows a good segmentation quality and a first cell-based quantitative analysis confirms the accordance with already known image-based quantitative results. A cell-based verification is subject of ongoing research.
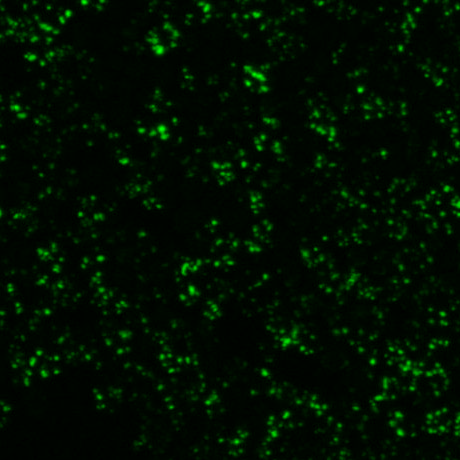 | 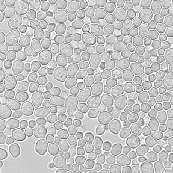 |  |
| fluorescence image (2D projection) | transmission image | labelled segmentation result |
 |  |  |  |  |
|
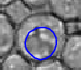
 |  | 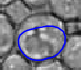 | 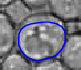 |  | 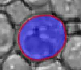 |
| The typical evolution of an active contour for cells with complex structure. | |||||
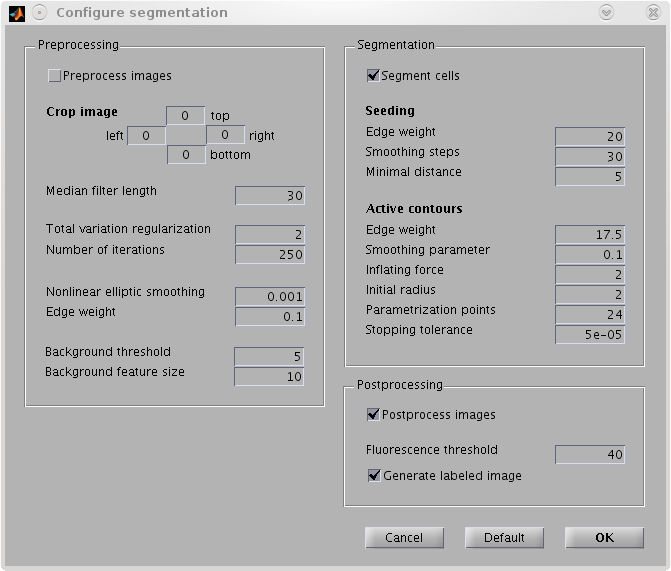 | 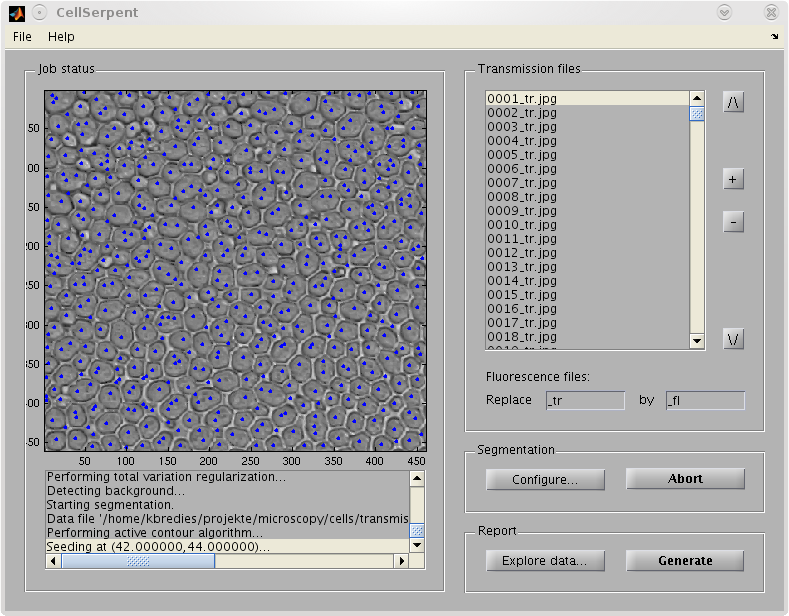 |
| GUI tool for biological applications. | |